Large Scale Learning
 Much of my current research focuses on kernel methods such as Support Vector Machines (SVMs) for sequence analysis problems appearing in bioinformatics. For example, I co-organized the PASCAL Large Scale Learning Challenge. Currently, we are preparing JMLR special topic on Large Scale Learning (soon to be published).
In addition, I am working on the design of new efficient string kernels as well
as faster algorithms to train and evaluate SVMs on sequences. Before I began to work on this topic, it had been almost unthinkable to train SVMs using sophisticated string kernels on more than a few hundred thousand examples. Using the newly developed methods we can now solve learning tasks involving up to 50 million training examples. Requiring reasonable amounts of computing time, we can now apply the resulting classifier to the whole human genome with as much as 6 billion examples.
Much of my current research focuses on kernel methods such as Support Vector Machines (SVMs) for sequence analysis problems appearing in bioinformatics. For example, I co-organized the PASCAL Large Scale Learning Challenge. Currently, we are preparing JMLR special topic on Large Scale Learning (soon to be published).
In addition, I am working on the design of new efficient string kernels as well
as faster algorithms to train and evaluate SVMs on sequences. Before I began to work on this topic, it had been almost unthinkable to train SVMs using sophisticated string kernels on more than a few hundred thousand examples. Using the newly developed methods we can now solve learning tasks involving up to 50 million training examples. Requiring reasonable amounts of computing time, we can now apply the resulting classifier to the whole human genome with as much as 6 billion examples.
Genomic Sequence Analysis
I have been working to employ these methods to splice site recognition
in several organisms (link). Together with my collaborators, I was able to show
that our methods drastically outperform all other methods, which is pivotal for the high accuracy of a novel
splice form prediction tool, mSplicer, and the
success of a related gene finding system, mGene, in the
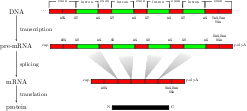 nGASP competition. Additionally, we have developed a promoter detection system
"ARTS" , that detects
transcription start sites on the whole human genome. Our approach works
with much higher accuracy than previous state of the art methods and by
using the developed large scale learning techniques, the SVMs could be
trained in only a few hours and applied genome wide.
nGASP competition. Additionally, we have developed a promoter detection system
"ARTS" , that detects
transcription start sites on the whole human genome. Our approach works
with much higher accuracy than previous state of the art methods and by
using the developed large scale learning techniques, the SVMs could be
trained in only a few hours and applied genome wide.
Interpretability
SVMs find a discrimination in a high dimensional kernel feature space
and as such often have to be treated as a black box. This implies that
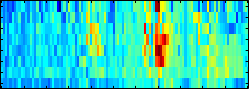 analyses or visualization of the learning result is inherently
difficult. It poses a problem for applications in bioinformatics as it
is often very important to understand which features are used for
learning and why the accuracy is high. I have developed a novel approach
based on Multiple
Kernel learning that
can be used for discovering discriminative features of the
underlying biological problem. An extended approach --- the so
called Positional Oligomer Importance Matrices (POIMs) ---
allows us to pin-point motifs, is very efficient and can be
directly applied to the learned SVM classifier.
analyses or visualization of the learning result is inherently
difficult. It poses a problem for applications in bioinformatics as it
is often very important to understand which features are used for
learning and why the accuracy is high. I have developed a novel approach
based on Multiple
Kernel learning that
can be used for discovering discriminative features of the
underlying biological problem. An extended approach --- the so
called Positional Oligomer Importance Matrices (POIMs) ---
allows us to pin-point motifs, is very efficient and can be
directly applied to the learned SVM classifier.
