Large Scale Learning with String Kernels
In genomic sequence analysis tasks like splice site recognition or promoter identification, large amounts of training sequences are available, and indeed needed to achieve sufficiently high classification performances. With my co-authors Gunnar Raetsch and Konrad Rieck I study string kernels that can be computed in linear time w.r.t. the length of the input sequences. In particular the recently proposed Spectrum kernel, the Weighted Degree kernel (WD) and the Weighted Degree kernel with shifts, which have been successfully used for various sequence analysis tasks.  We discuss extensions using data structures such as tries and suffix trees as well as modifications of a SVM chunking algorithm in order to significantly accelerate SVM training and their evaluation on test sequences. Our simulations using the WD kernel and Spectrum kernel show that large scale SVM training can be accelerated by factors of 7 and 60 times, respectively, while requiring considerably less memory. We demonstrate that these algorithms can be effectively parallelized for further acceleration. Our method allows us to train SVMs on sets as large as 10 million sequences and solve Multiple Kernel Learning problems with 1 million sequences. Moreover, using these techniques the evaluation on new sequences is often several thousand times faster, allowing us to apply the classifiers on genome-sized data sets with seven billion test examples. We finally demonstrate how the proposed data structures can be used to understand the SVM classifiers decision function. All presented algorithms are implemented in our Machine Learning toolbox SHOGUN for which the source code is publicly available at http://www.shogun-toolbox.org.
We discuss extensions using data structures such as tries and suffix trees as well as modifications of a SVM chunking algorithm in order to significantly accelerate SVM training and their evaluation on test sequences. Our simulations using the WD kernel and Spectrum kernel show that large scale SVM training can be accelerated by factors of 7 and 60 times, respectively, while requiring considerably less memory. We demonstrate that these algorithms can be effectively parallelized for further acceleration. Our method allows us to train SVMs on sets as large as 10 million sequences and solve Multiple Kernel Learning problems with 1 million sequences. Moreover, using these techniques the evaluation on new sequences is often several thousand times faster, allowing us to apply the classifiers on genome-sized data sets with seven billion test examples. We finally demonstrate how the proposed data structures can be used to understand the SVM classifiers decision function. All presented algorithms are implemented in our Machine Learning toolbox SHOGUN for which the source code is publicly available at http://www.shogun-toolbox.org.
Large Scale Learning Challenge
 This PASCAL Challenge is concerned with the scalability and
efficiency of existing ML approaches with respect to
computational, memory or communication resources, e.g.
resulting from a high algorithmic complexity, from the size or
dimensionality of the data set, and from the trade-off between
distributed resolution and communication costs.
This PASCAL Challenge is concerned with the scalability and
efficiency of existing ML approaches with respect to
computational, memory or communication resources, e.g.
resulting from a high algorithmic complexity, from the size or
dimensionality of the data set, and from the trade-off between
distributed resolution and communication costs.
This competition is designed to be fair and enables a direct comparison of current large scale classifiers aimed at answering the question “Which learning method is the most accurate given limited resources?”. To this end we provide a generic evaluation framework tailored to the specifics of the competing methods. Providing a wide range of datasets, each of which having specific properties we propose to evaluate the methods based on performance figures, displaying training time vs. test error, dataset size vs. test error and dataset size vs. training time.
While the challenge is over, we still allow submissions to the challenge using the submission system. In addition, you may browse through the results and render result figures adjusted to your needs.
Currently, we are preparing JMLR special topic on Large Scale Learning (soon to be published).
I wrote and designed this submission system from scratch using python and django.
Optimized Cutting Plane Algorithm
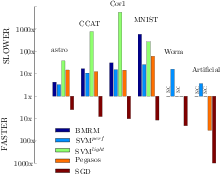 Together with Vojtech Franc I have developed an optimized cutting plane algorithm (OCA) for solving large scale risk minimization problems. We prove that the number of iterations OCA requires to converge to a epsilon-precise solution is approximately linear in the sample size. We derive OCAS, an OCA based linear binary Support Vector Machine (SVM) solver, and OCAM, a linear multi-class SVM solver. In an extensive empirical evaluation we show that OCAS outperforms current state of the art SVM solvers, like SVMlight, SVMperf and BMRM, achieving speedups of over 1,200 on some datasets over SVMlight and 29 over SVMperf, while obtaining the same precise Support Vector solution. OCAS even in the early optimization steps shows often faster convergence than the so far in this domain prevailing approximative methods SGD and Pegasos. In addition, our proposed linear multi-class SVM solver OCAM achieves speedups of up to 10 compared to SVMmulti-class . Finally, we apply OCAS and OCAM to two real-world applications, the problem of human acceptor splice site detection and malware detection. Effectively parallelizing OCAS we achieve state-of-the art results on the acceptor splice site recognition problem only by being able to learn on all the available 50 million examples in a 12 million dimensional feature space.
Together with Vojtech Franc I have developed an optimized cutting plane algorithm (OCA) for solving large scale risk minimization problems. We prove that the number of iterations OCA requires to converge to a epsilon-precise solution is approximately linear in the sample size. We derive OCAS, an OCA based linear binary Support Vector Machine (SVM) solver, and OCAM, a linear multi-class SVM solver. In an extensive empirical evaluation we show that OCAS outperforms current state of the art SVM solvers, like SVMlight, SVMperf and BMRM, achieving speedups of over 1,200 on some datasets over SVMlight and 29 over SVMperf, while obtaining the same precise Support Vector solution. OCAS even in the early optimization steps shows often faster convergence than the so far in this domain prevailing approximative methods SGD and Pegasos. In addition, our proposed linear multi-class SVM solver OCAM achieves speedups of up to 10 compared to SVMmulti-class . Finally, we apply OCAS and OCAM to two real-world applications, the problem of human acceptor splice site detection and malware detection. Effectively parallelizing OCAS we achieve state-of-the art results on the acceptor splice site recognition problem only by being able to learn on all the available 50 million examples in a 12 million dimensional feature space.
Large Scale Multiple Kernel Learning
While classical kernel-based learning algorithms are based on a single kernel, in practice it is often desirable to use multiple kernels. Lankriet et al. (2004) considered conic combinations of kernel matrices for classification, leading to a convex quadratically constrained quadratic program. We show that it can be rewritten as a semi-infinite linear program that can be efficiently solved by recycling the standard SVM implementations. Moreover, we generalize the formulation and our method to a larger class of problems, including regression and one-class classification. Experimental results show that the proposed algorithm works for hundred thousands of examples or hundreds of kernels to be combined, and helps for automatic model selection, improving the interpretability of the learning result. In a second part we discuss general speed up mechanism for SVMs, especially when used with sparse feature maps as appear for string kernels, allowing us to train a string kernel SVM on a 10 million real-world splice dataset from computational biology. We integrated Multiple Kernel Learning in our Machine Learning toolbox SHOGUN for which the source code is publicly available at http://www.shogun-toolbox.org.
COFFIN
In a variety of applications, kernel machines such as Support Vector Machines (SVMs) have been used with great success often delivering state-of-the-art results. Using the kernel trick, they work on several domains and even enable heterogeneous data fusion by concatenating feature spaces or multiple kernel learning. Unfortunately, they are not suited for truly large-scale applications since they suffer from the curse of supporting vectors, e.g., the speed of applying SVMs decays linearly with the number of support vectors. In this paper we develop COFFIN --- a new training strategy for linear SVMs that effectively allows the use of on demand computed kernel feature spaces and virtual examples in the primal. With linear training and prediction effort this framework leverages SVM applications to truly large-scale problems: As an example, we train SVMs for human splice site recognition involving 50 million examples and sophisticated string kernels. Additionally, we learn an SVM based gender detector on 5 million examples on low-tech hardware and achieve beyond the state-of-the-art accuracies on both tasks. Click more to access source code, data sets and scripts.
